Prof. Jun Liu’s team revealed the characteristics and molecular mechanisms of the T cell immune escape for SARS-CoV-2
Source:Jun Liu
2025-02-24
New variants of SARS-CoV-2 are continuously emerging with mutations that enable escape from antibody responses. However, it is unclear whether these variants also affect T cell immunity, in particular the recent variants such as BA.2.86 and its sub-variant JN.1, which is now a predominant global variant of interest. Some studies show that memory T cells can still recognize BA.2.86 in many cases, but there is also evidence of immune escape in certain epitopes of these variants and sub-variants. However, we do not yet have enough evidence to determine how mutations in BA.2.86 and JN.1 affect T cell recognition of specific HLA-restricted epitopes. It is crucial to study how these mutations allow the virus to evade T cell immunity, as this could help us to understand the ongoing evolution of SARS-CoV-2.
On January 28, 2025, a study entitled ‘T cell immunity evasion by SARS-CoV-2 JN.1 escapees targeting two cytotoxic T cell epitope hotspots’ was published in Nature Immunology, led by Professor Jun Liu, Academician George F. Gao and Associate Professor Yingze Zhao from Chinese Center for Disease Control and Prevention, in collaboration with Professor Jikun Zhou from the Fifth Hospital of Shijiazhuang and Professor Rui Song from the Beijing Ditan Hospital. This study describes how newly emerging mutations in JN.1 contribute to evasion of CD8+ T cell responses. Mutations occurring in T cell epitope hotspots in the viral spike protein and in a highly conserved site in the viral nucleocapsid suggest that T cell-mediated immune pressure is a key driving force for SARS-CoV-2 evolution and adaptation.
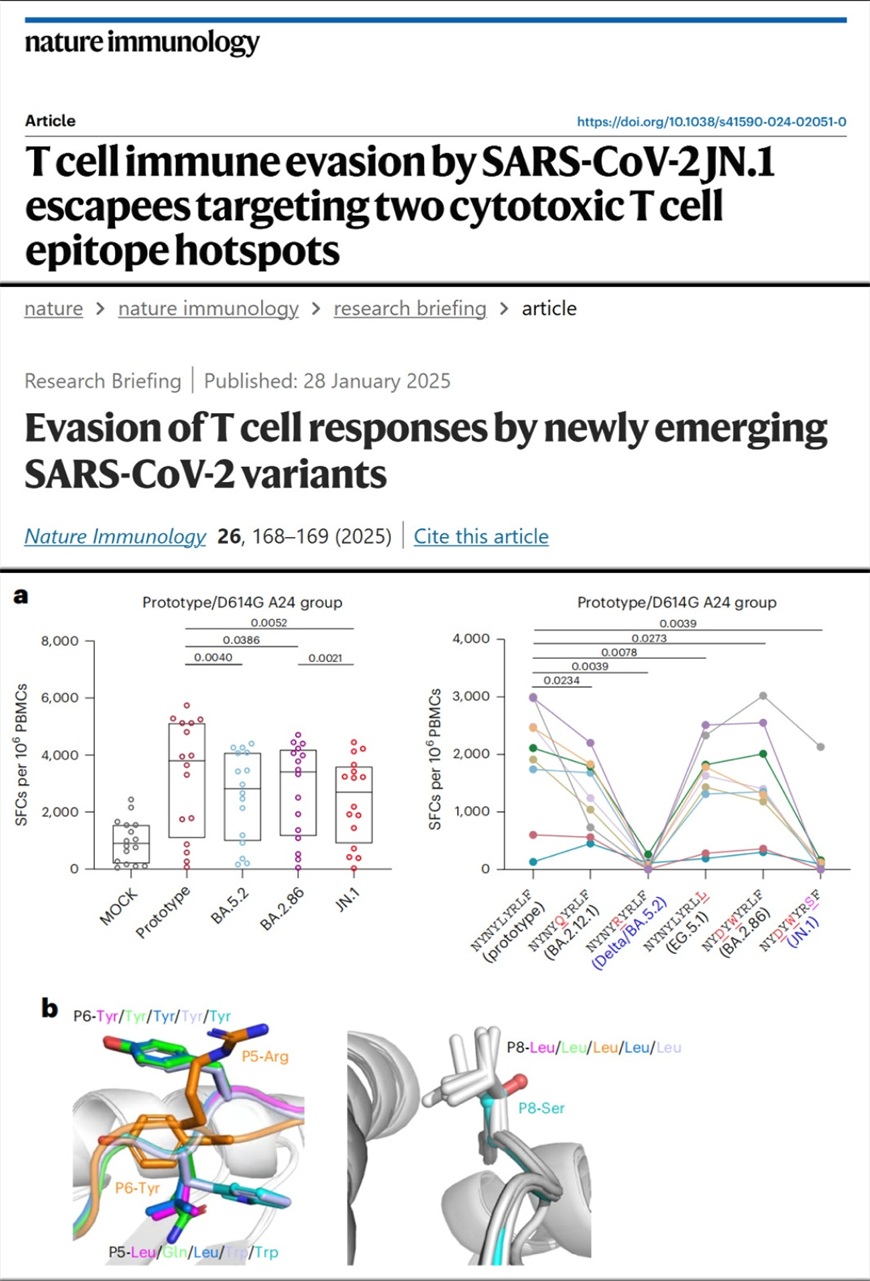
To investigated how mutations in JN.1 affect T cell immunity, the team synthesized the overlapping long peptides and HLA class I-restricted peptides covering characteristic mutation sites in the viral proteins spike, membrane, nucleocapsid and open reading frame 1ab-encoded proteins. Using a highly sensitive adjusted ELISpot-based T cell assay they have proposed before, the team tested the T cell responses to these peptides and determined which mutations may enable immune escape. To validate these findings, they further examined T cell responses in blood samples from people who had recovered from infections with different SARS-CoV-2 strains. The results showed a correlation between the specific HLA type and T cell immune escape and led to the identification of two epitope hotspots. They found that although T cell cross-recognition of the S1 peptide pools was present, cross CD8+ T cell responses to BA.5.2 and JN.1 among HLA-A*2402-positive populations were reduced compared with responses to other sub-variants.
Moreover, the analysis highlighted the epitope HLA-A*2402–S448–456 as a mutation hotspot. The team tested a series of mutations in this epitope as found in different viral sub-variants and observed a sharp reduction in CD8+ T cell responses for mutations, L452R in Delta/BA.5.2 and N450D/L452W/L455S in JN.1. The team also explored how the mutations affect HLA binding, T cell receptor docking, the overall conformation of the peptide and how the peptides fit in the HLA binding groove. Structural analysis showed that changes in peptide hydrophobicity due to mutations such as L452R and L455S had a key role in immune evasion, by affecting T cell receptor anchoring. In addition, the team also identified that Q229 in nucleocapsid protein is conserved between SARS-CoV, SARS-CoV-2 and most other Sarbecoviruses, mutation of which, first emerging in BA.2.86 and JN.1, can contribute to immune evasion in people with HLA-A2-related haplotype.
This study reveals how the JN.1 sub-variant evades T cell immunity and sheds lights on why certain variants become dominant. It highlights the role of T cell-mediated immune pressure in driving new mutations in the virus. Mutations at T cell epitope hotspots may influence the ability of the global population to defend against the virus in an HLA allele-restricted manner. The importance of T cell immunity in the surveillance and characterization of SARS-CoV-2 variants and sub-variants, and in the development of universal vaccines must also be carefully considered.
Professor Jun Liu from Chinese Center for Disease Control and Prevention served as the leading corresponding author, with Academician George F. Gao and Associate Professor Yingze Zhao, in collaboration with Professor Jikun Zhou from the Fifth Hospital of Shijiazhuang and Professor Rui Song from the Beijing Ditan Hospital as co-corresponding authors. The first authors of the study are the PhD students in the team: Jinmin Tian, Bingli Shang (Wenzhou Medical University), Jianing Zhang, Yuanyuan Guo (Shandong University), Min Li and Yuechao Hu.
Article Link: https://doi.org/10.1038/s41590-024-02051-0
On January 28, 2025, a study entitled ‘T cell immunity evasion by SARS-CoV-2 JN.1 escapees targeting two cytotoxic T cell epitope hotspots’ was published in Nature Immunology, led by Professor Jun Liu, Academician George F. Gao and Associate Professor Yingze Zhao from Chinese Center for Disease Control and Prevention, in collaboration with Professor Jikun Zhou from the Fifth Hospital of Shijiazhuang and Professor Rui Song from the Beijing Ditan Hospital. This study describes how newly emerging mutations in JN.1 contribute to evasion of CD8+ T cell responses. Mutations occurring in T cell epitope hotspots in the viral spike protein and in a highly conserved site in the viral nucleocapsid suggest that T cell-mediated immune pressure is a key driving force for SARS-CoV-2 evolution and adaptation.

To investigated how mutations in JN.1 affect T cell immunity, the team synthesized the overlapping long peptides and HLA class I-restricted peptides covering characteristic mutation sites in the viral proteins spike, membrane, nucleocapsid and open reading frame 1ab-encoded proteins. Using a highly sensitive adjusted ELISpot-based T cell assay they have proposed before, the team tested the T cell responses to these peptides and determined which mutations may enable immune escape. To validate these findings, they further examined T cell responses in blood samples from people who had recovered from infections with different SARS-CoV-2 strains. The results showed a correlation between the specific HLA type and T cell immune escape and led to the identification of two epitope hotspots. They found that although T cell cross-recognition of the S1 peptide pools was present, cross CD8+ T cell responses to BA.5.2 and JN.1 among HLA-A*2402-positive populations were reduced compared with responses to other sub-variants.
Moreover, the analysis highlighted the epitope HLA-A*2402–S448–456 as a mutation hotspot. The team tested a series of mutations in this epitope as found in different viral sub-variants and observed a sharp reduction in CD8+ T cell responses for mutations, L452R in Delta/BA.5.2 and N450D/L452W/L455S in JN.1. The team also explored how the mutations affect HLA binding, T cell receptor docking, the overall conformation of the peptide and how the peptides fit in the HLA binding groove. Structural analysis showed that changes in peptide hydrophobicity due to mutations such as L452R and L455S had a key role in immune evasion, by affecting T cell receptor anchoring. In addition, the team also identified that Q229 in nucleocapsid protein is conserved between SARS-CoV, SARS-CoV-2 and most other Sarbecoviruses, mutation of which, first emerging in BA.2.86 and JN.1, can contribute to immune evasion in people with HLA-A2-related haplotype.
This study reveals how the JN.1 sub-variant evades T cell immunity and sheds lights on why certain variants become dominant. It highlights the role of T cell-mediated immune pressure in driving new mutations in the virus. Mutations at T cell epitope hotspots may influence the ability of the global population to defend against the virus in an HLA allele-restricted manner. The importance of T cell immunity in the surveillance and characterization of SARS-CoV-2 variants and sub-variants, and in the development of universal vaccines must also be carefully considered.
Professor Jun Liu from Chinese Center for Disease Control and Prevention served as the leading corresponding author, with Academician George F. Gao and Associate Professor Yingze Zhao, in collaboration with Professor Jikun Zhou from the Fifth Hospital of Shijiazhuang and Professor Rui Song from the Beijing Ditan Hospital as co-corresponding authors. The first authors of the study are the PhD students in the team: Jinmin Tian, Bingli Shang (Wenzhou Medical University), Jianing Zhang, Yuanyuan Guo (Shandong University), Min Li and Yuechao Hu.
Article Link: https://doi.org/10.1038/s41590-024-02051-0


