Prof. Linqi Zhang and Zheng Zhang et al revealed the molecular mechanism of broadly neutralizing antibodies elicited by wild-type SARS-CoV-2 infection
Source:Bin Ju, Zheng Zhang
2023-03-24
On March 13, 2023, a team of Prof. Zheng Zhang and Dr. Bin Ju from Shenzhen Third People's Hospital (The Second Affiliated Hospital of Southern University of Science and Technology)/Shenzhen National Clinical Research Center for Infectious Diseases, in collaboration with Prof. Linqi Zhang, Prof. Xinquan Wang from Tsinghua University, and Prof. Justin Jang Hann Chu from National University of Singapore, published a research article entitled "Infection with wild-type SARS-CoV-2 elicits broadly neutralizing and protective antibodies against omicron subvariants" in Nature Immunology. This is another important work of the collaborative team in the research of anti-SARS-CoV-2 antibodies, providing some important scientific basis and guidance for the optimization of antibody drugs and vaccines.
The rapid emergence and turnover of various SARS-CoV-2 variants have posed considerable challenges to the infection- and vaccination-elicited immune barrier. Omicron subvariants are the most typical representatives of these SARS-CoV-2 variants and the most efficient in transmission around the world. The exceptionally high number of mutations in the spike (S) protein of omicron has resulted in their strong capabilities to escape from the recognition of antibodies. The major questions that remain to be answered are the extent of such escape and whether antibodies in the infected or vaccinated individuals can neutralize these variants identified so far. If such neutralizing antibodies (nAbs) do exist, it is important to identify their binding epitopes, antiviral mechanisms, and potential protective capacities against SARS-CoV-2 variants. Answering these questions is essential for helping us to understand the human antibody response and will provide critical guides for the optimization of next-generation vaccines and immunization strategies.
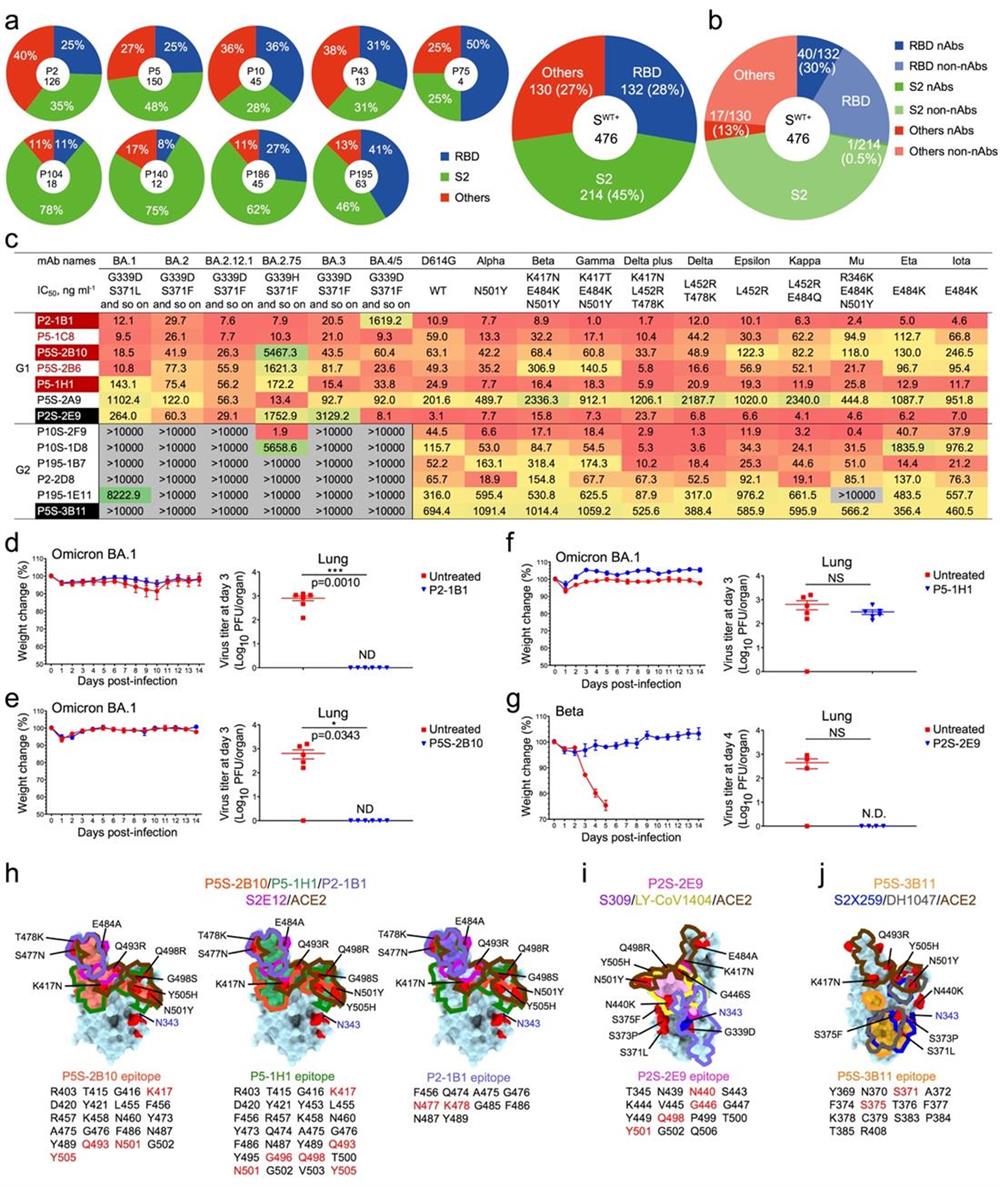
In their study, the researchers isolated a total of 476 S-specific monoclonal antibodies (mAbs) from nine convalescent COVID-19 patients infected with the wild-type SARS-CoV-2. Among these 476 mAbs, 45% were S2-specific, 28% were receptor-binding domain (RBD)-specific, and 27% targeted other epitopes (Fig. 1a). However, pseudovirus-based neutralization assay revealed that the majority of nAbs were RBD-specific (30%, 40/132), followed by NTD-specific mAbs (17/130) and S2-specific mAbs (1/214) (Fig. 1b), suggesting that RBD is the critical antigenic component responsible for inducing neutralizing antibodies. Then, the researchers evaluated the broadly neutralizing activities of 40 RBD-specific mAbs. Among them, 13 mAbs could maintain effective neutralization against some concerned SARS-CoV-2 variants, such as beta and delta, etc. However, with the emergence of the omicron variants, 6 of these antibodies lost their neutralizing activities. Surprisingly, the rest of 7 mAbs still neutralized all tested variants, indicating that the immune imprint generated by the wild-type SARS-CoV-2 infection has the potential to combat the antibody evasion of the following emerged SARS-CoV-2 variants. Further genetic analysis showed that these potent broad nAbs (bnAbs) predominantly utilized the IGHV3-53 gene family, highlighting the importance of these "public antibodies" once again (Fig. 1c). Animal protection experiments showed that four representative bnAbs (P2-1B1, P5S-2B10, P5-1H1, P2S-2E9) effectively inhibited the infection of omicron BA.1 and beta variants (Fig. 1d-g). Structural analysis revealed that these bnAbs recognized diverse epitopes on the RBD and could tolerate a certain degree of mutations at their binding sites. P5S-2B10, P5-1H1, and P2-1B1 belonged to the typical Class 1 antibodies, mimicking and directly competing with the ACE2 receptor. P2S-2E9 was a Class 3 antibody, and P5S-3B11 was a Class 4 antibody (Fig. 1h-j). Overall, this study conclusively answered three critical scientific questions: 1) Infection with the wild-type SARS-CoV-2 can elicit potent bnAbs; 2) The RBD protein is still the ideal target for inducing effective bnAbs; 3) Some nAbs that mimicking receptor binding and favoring the usage of IGHV3-53 gene family play important roles in fighting against SARS-CoV-2 variants.
Further research results (unpublished data) demonstrated that some of these bnAbs could maintain effective neutralizing activities against BF.7 and BA.5.2 (circulating in China), as well as BQ.1, BQ.1.1, XBB, XBB.1, and XBB.1.5, which are spreading globally. This study not only offers us a comprehensive understanding of human antibody response, but also provides crucial guidance for the design and optimization of next-generation SARS-CoV-2 vaccines. While these isolated bnAbs could serve as promising candidates for the development of novel antibody therapies, a deeper understanding of how the memory B cells producing these antibodies are initially induced, maintained, and recalled will hold the key to rational design of vaccines capable of selectively boosting such desirable antibodies against SARS-CoV-2 variants and beyond.
Prof. Linqi Zhang from Tsinghua University, Prof. Zheng Zhang from Shenzhen Third People's Hospital (The Second Affiliated Hospital of Southern University of Science and Technology)/Shenzhen National Clinical Research Center for Infectious Diseases, Prof. Xinquan Wang from Tsinghua University, and Prof. Justin Jang Hann Chu from National University of Singapore are co-corresponding authors. Dr. Bin Ju from Shenzhen Third People's Hospital, Dr. Qi Zhang from Tsinghua University, PhD candidate Ziyi Wang from Tsinghua University, Dr. Zhen Qin Aw from National University of Singapore, and PhD candidate Peng Chen from Tsinghua University are co-first authors. This study was supported by National Key Plan for Scientific Research and Development of China, National Natural Science Foundation, Beijing Municipal Science and Technology Commission, Guangdong Basic and Applied Basic Research Foundation, Shenzhen Science and Technology Program, Wanke Scientific Research Program, Chunfeng Foundation, Tencent Foundation, Shuidi Foundation, TH Capital, and others.
Article link: https://www.nature.com/articles/s41590-023-01449-6

The rapid emergence and turnover of various SARS-CoV-2 variants have posed considerable challenges to the infection- and vaccination-elicited immune barrier. Omicron subvariants are the most typical representatives of these SARS-CoV-2 variants and the most efficient in transmission around the world. The exceptionally high number of mutations in the spike (S) protein of omicron has resulted in their strong capabilities to escape from the recognition of antibodies. The major questions that remain to be answered are the extent of such escape and whether antibodies in the infected or vaccinated individuals can neutralize these variants identified so far. If such neutralizing antibodies (nAbs) do exist, it is important to identify their binding epitopes, antiviral mechanisms, and potential protective capacities against SARS-CoV-2 variants. Answering these questions is essential for helping us to understand the human antibody response and will provide critical guides for the optimization of next-generation vaccines and immunization strategies.
In their study, the researchers isolated a total of 476 S-specific monoclonal antibodies (mAbs) from nine convalescent COVID-19 patients infected with the wild-type SARS-CoV-2. Among these 476 mAbs, 45% were S2-specific, 28% were receptor-binding domain (RBD)-specific, and 27% targeted other epitopes (Fig. 1a). However, pseudovirus-based neutralization assay revealed that the majority of nAbs were RBD-specific (30%, 40/132), followed by NTD-specific mAbs (17/130) and S2-specific mAbs (1/214) (Fig. 1b), suggesting that RBD is the critical antigenic component responsible for inducing neutralizing antibodies. Then, the researchers evaluated the broadly neutralizing activities of 40 RBD-specific mAbs. Among them, 13 mAbs could maintain effective neutralization against some concerned SARS-CoV-2 variants, such as beta and delta, etc. However, with the emergence of the omicron variants, 6 of these antibodies lost their neutralizing activities. Surprisingly, the rest of 7 mAbs still neutralized all tested variants, indicating that the immune imprint generated by the wild-type SARS-CoV-2 infection has the potential to combat the antibody evasion of the following emerged SARS-CoV-2 variants. Further genetic analysis showed that these potent broad nAbs (bnAbs) predominantly utilized the IGHV3-53 gene family, highlighting the importance of these "public antibodies" once again (Fig. 1c). Animal protection experiments showed that four representative bnAbs (P2-1B1, P5S-2B10, P5-1H1, P2S-2E9) effectively inhibited the infection of omicron BA.1 and beta variants (Fig. 1d-g). Structural analysis revealed that these bnAbs recognized diverse epitopes on the RBD and could tolerate a certain degree of mutations at their binding sites. P5S-2B10, P5-1H1, and P2-1B1 belonged to the typical Class 1 antibodies, mimicking and directly competing with the ACE2 receptor. P2S-2E9 was a Class 3 antibody, and P5S-3B11 was a Class 4 antibody (Fig. 1h-j). Overall, this study conclusively answered three critical scientific questions: 1) Infection with the wild-type SARS-CoV-2 can elicit potent bnAbs; 2) The RBD protein is still the ideal target for inducing effective bnAbs; 3) Some nAbs that mimicking receptor binding and favoring the usage of IGHV3-53 gene family play important roles in fighting against SARS-CoV-2 variants.
Further research results (unpublished data) demonstrated that some of these bnAbs could maintain effective neutralizing activities against BF.7 and BA.5.2 (circulating in China), as well as BQ.1, BQ.1.1, XBB, XBB.1, and XBB.1.5, which are spreading globally. This study not only offers us a comprehensive understanding of human antibody response, but also provides crucial guidance for the design and optimization of next-generation SARS-CoV-2 vaccines. While these isolated bnAbs could serve as promising candidates for the development of novel antibody therapies, a deeper understanding of how the memory B cells producing these antibodies are initially induced, maintained, and recalled will hold the key to rational design of vaccines capable of selectively boosting such desirable antibodies against SARS-CoV-2 variants and beyond.

Fig. 1 Infection with wild-type SARS-CoV-2 elicits broadly neutralizing and protective antibodies against omicron subvariants.
Prof. Linqi Zhang from Tsinghua University, Prof. Zheng Zhang from Shenzhen Third People's Hospital (The Second Affiliated Hospital of Southern University of Science and Technology)/Shenzhen National Clinical Research Center for Infectious Diseases, Prof. Xinquan Wang from Tsinghua University, and Prof. Justin Jang Hann Chu from National University of Singapore are co-corresponding authors. Dr. Bin Ju from Shenzhen Third People's Hospital, Dr. Qi Zhang from Tsinghua University, PhD candidate Ziyi Wang from Tsinghua University, Dr. Zhen Qin Aw from National University of Singapore, and PhD candidate Peng Chen from Tsinghua University are co-first authors. This study was supported by National Key Plan for Scientific Research and Development of China, National Natural Science Foundation, Beijing Municipal Science and Technology Commission, Guangdong Basic and Applied Basic Research Foundation, Shenzhen Science and Technology Program, Wanke Scientific Research Program, Chunfeng Foundation, Tencent Foundation, Shuidi Foundation, TH Capital, and others.
Article link: https://www.nature.com/articles/s41590-023-01449-6


