Prof. Ning Zhang, Zemin Zhang and Jiye Zhu’s team reported liver tumor microenvironment subtypes and neutrophil heterogeneity
Source:Ruidong Xue
2022-12-28
Recently, team of Professor Ning Zhang from Translational Cancer Research Center of Peking University First Hospital, in collaboration with Professor Zemin Zhang from Biomedical Pioneering Innovation Center (BIOPIC) of Peking University, and Professor Jiye Zhu from Department of Hepatobiliary Surgery of Peking University People's Hospital performed a comprehensive single-cell analysis of primary liver cancer. They defined the TIMELASER subtypes for liver cancer immune microenvironment at the single cell resolution, revealed the phenotypic and functional heterogeneity of tumor associated neutrophils (TANs), and showed that targeting TAN is a promising new immunotherapy regimen for treating liver cancer. This study, titled "Liver tumour immune microenvironment subtypes and neutrophil heterogeneity," was published in Nature on November 9, 2022.
The heterogeneity of tumor immune microenvironment (TIME), organized by various immune and stromal cells, is a major cause of tumor metastasis and drug resistance, but how different TIME subtypes are connected to the clinical relevance in liver cancer remains elusive. Recent clinical trials of immune checkpoint blockade and related combination therapies have shown promising results that would change the PLC therapeutic paradigm. However, previous single-cell studies usually involve antibody-based enrichment of certain cell types and are limited in cohort size. Therefore, a comprehensive single cell study covering most cell populations and three major subtypes of PLC with established clinical parameters is needed.
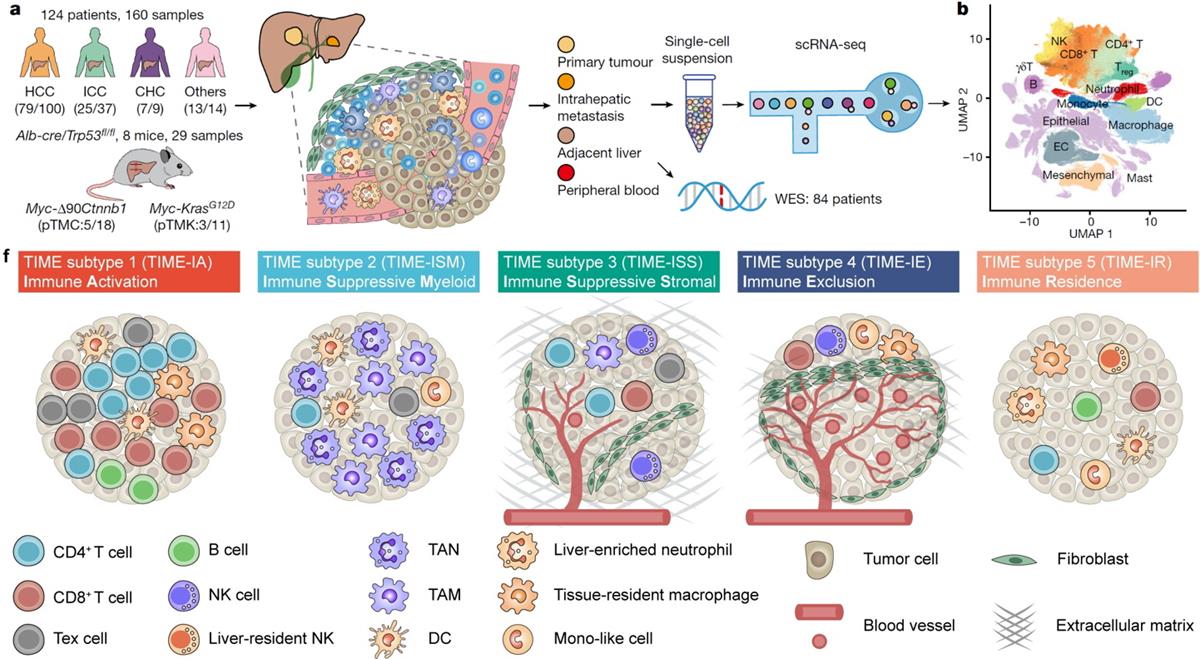
To systematically survey the TIME landscape across liver cancer covering all cell populations, a total of 189 samples from human and mouse were analyzed by single-cell RNA-seq. Based on the differential enrichment of 89 TIME cell subpopulations identified in our dataset, five different TIME subtypes were identified, including ① Immune Activation (TIME-IA); ② Immune Suppressive Myeloid (TIME-ISM); ③ Immune Suppressive Stromal (TIME-ISS); ④Immune Exclusion (TIME-IE); ⑤ Immune Residence (TIME-IR), together named the TIMELASER framework. Our TIMELASER framework can be successfully validated by published liver cancer datasets based on bulk RNA-seq, scRNA-seq, spatial transcriptome, and multiplex CODEX staining data. The authors also observe cancer type preferences of TIMELASER subtypes in PLC, underscoring technical advantages of single cell analysis for TIME landscape and indicating different therapeutic strategies for different PLC subtypes.
Neutrophils are very fragile and can survive no more than a week after entering the peripheral blood and no more than 24 hours outside the body. Therefore, most previous single-cell studies of liver cancer failed to capture neutrophils. In this study, largely owing to the rapid and antibody-free experimental strategy, more than 30,000 neutrophils were successfully captured and divided into 11 neutrophil subsets, among which 6 subsets of TANs were identified. The authors analyzed the developmental trajectory and key transcription factors of these neutrophil subsets, and found that two neutrophil subsets, CCL4+ TAN and PD-L1+ TAN, may play potential pro-tumor roles. Further, they constructed in vitro tumor cell line-neutrophil co-culture system and also performed ex vivo analysis of patient-derived neutrophil. They showed that CCL4+ TANs could promote tumor growth by recruiting tumor-related macrophages and that PD-L1+ TAN promote tumor growth by inhibiting cytotoxic CD8+ T cells. To further explore the function of TANs in vivo, the authors characterized neutrophils in two spontaneous liver cancer mouse models, and with the anti-Ly6G antibody, they showed that neutrophil depletion could attenuate tumor progression. Further analysis showed that depletion of neutrophils could alter the TAN composition in mouse models. Targeting multiple pro-tumor TAN subsets and elevating the proportion of Apoa2+ TANs might both serve as promising immunotherapies for liver cancer. Further exploring the impact of neutrophils on immune checkpoint blockade and related confounding clinical factors would offer new opportunities to better understand TAN biology and propose translational research avenues for treating liver cancer.
Dr. Ruidong Xue from Peking University First Hospital, Dr. Qiming Zhang from BIOPIC, Qi Cao from Peking University First Hospital, Dr. Ruirui Kong from Peking University First Hospital and Dr. Xiao Xiang from Peking University People's Hospital are co-first authors. This project was jointly supported by multiple fundings from the National Science Foundation of China (including the Basic Science Center Program 81988101, 82173035, 81802813,82030079, 81872508, 81972656, 81972735, and 81902401) and the National Science and Technology Major Project of China (2018ZX10723204), the Michigan Medicine and Peking University Health Science Center Joint Institute for Translational and Clinical Research (BMU2020JI005).
Article link:https://www.nature.com/articles/s41586-022-05400-x
The heterogeneity of tumor immune microenvironment (TIME), organized by various immune and stromal cells, is a major cause of tumor metastasis and drug resistance, but how different TIME subtypes are connected to the clinical relevance in liver cancer remains elusive. Recent clinical trials of immune checkpoint blockade and related combination therapies have shown promising results that would change the PLC therapeutic paradigm. However, previous single-cell studies usually involve antibody-based enrichment of certain cell types and are limited in cohort size. Therefore, a comprehensive single cell study covering most cell populations and three major subtypes of PLC with established clinical parameters is needed.
To systematically survey the TIME landscape across liver cancer covering all cell populations, a total of 189 samples from human and mouse were analyzed by single-cell RNA-seq. Based on the differential enrichment of 89 TIME cell subpopulations identified in our dataset, five different TIME subtypes were identified, including ① Immune Activation (TIME-IA); ② Immune Suppressive Myeloid (TIME-ISM); ③ Immune Suppressive Stromal (TIME-ISS); ④Immune Exclusion (TIME-IE); ⑤ Immune Residence (TIME-IR), together named the TIMELASER framework. Our TIMELASER framework can be successfully validated by published liver cancer datasets based on bulk RNA-seq, scRNA-seq, spatial transcriptome, and multiplex CODEX staining data. The authors also observe cancer type preferences of TIMELASER subtypes in PLC, underscoring technical advantages of single cell analysis for TIME landscape and indicating different therapeutic strategies for different PLC subtypes.
Neutrophils are very fragile and can survive no more than a week after entering the peripheral blood and no more than 24 hours outside the body. Therefore, most previous single-cell studies of liver cancer failed to capture neutrophils. In this study, largely owing to the rapid and antibody-free experimental strategy, more than 30,000 neutrophils were successfully captured and divided into 11 neutrophil subsets, among which 6 subsets of TANs were identified. The authors analyzed the developmental trajectory and key transcription factors of these neutrophil subsets, and found that two neutrophil subsets, CCL4+ TAN and PD-L1+ TAN, may play potential pro-tumor roles. Further, they constructed in vitro tumor cell line-neutrophil co-culture system and also performed ex vivo analysis of patient-derived neutrophil. They showed that CCL4+ TANs could promote tumor growth by recruiting tumor-related macrophages and that PD-L1+ TAN promote tumor growth by inhibiting cytotoxic CD8+ T cells. To further explore the function of TANs in vivo, the authors characterized neutrophils in two spontaneous liver cancer mouse models, and with the anti-Ly6G antibody, they showed that neutrophil depletion could attenuate tumor progression. Further analysis showed that depletion of neutrophils could alter the TAN composition in mouse models. Targeting multiple pro-tumor TAN subsets and elevating the proportion of Apoa2+ TANs might both serve as promising immunotherapies for liver cancer. Further exploring the impact of neutrophils on immune checkpoint blockade and related confounding clinical factors would offer new opportunities to better understand TAN biology and propose translational research avenues for treating liver cancer.
Dr. Ruidong Xue from Peking University First Hospital, Dr. Qiming Zhang from BIOPIC, Qi Cao from Peking University First Hospital, Dr. Ruirui Kong from Peking University First Hospital and Dr. Xiao Xiang from Peking University People's Hospital are co-first authors. This project was jointly supported by multiple fundings from the National Science Foundation of China (including the Basic Science Center Program 81988101, 82173035, 81802813,82030079, 81872508, 81972656, 81972735, and 81902401) and the National Science and Technology Major Project of China (2018ZX10723204), the Michigan Medicine and Peking University Health Science Center Joint Institute for Translational and Clinical Research (BMU2020JI005).

Figure. Research schematic of TIMELASER subtypes and neutrophil heterogeneity
Article link:https://www.nature.com/articles/s41586-022-05400-x


